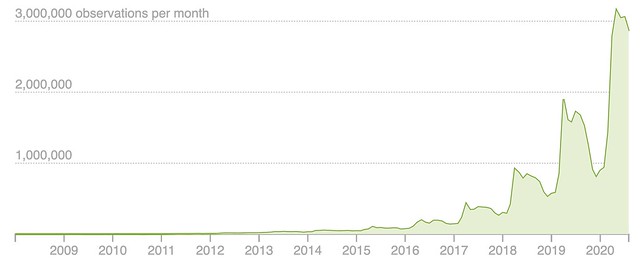
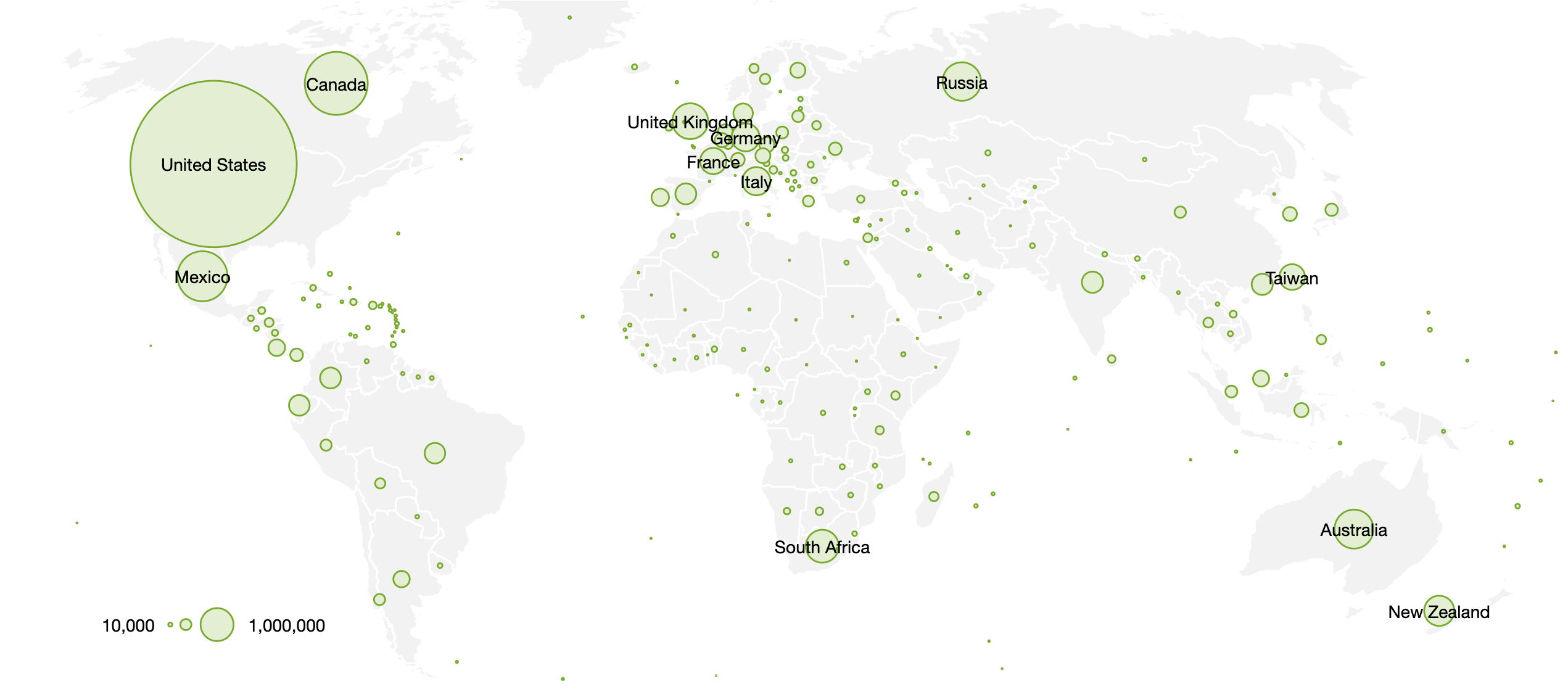
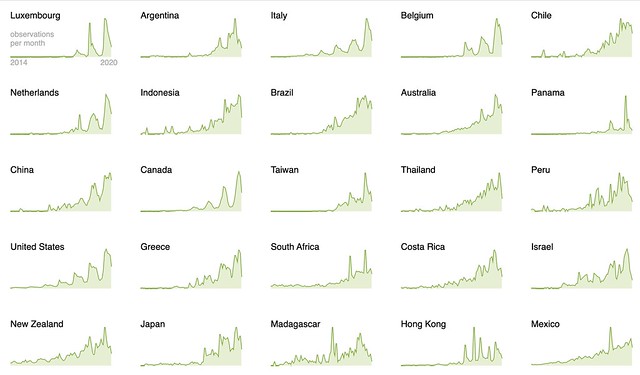
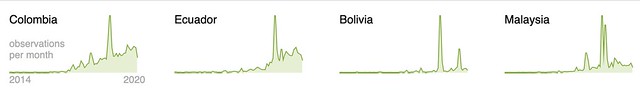
We’ve introduced a new way to use Taxon Splits that gives curators more flexibility. Here’s some background before explaining the change. If this background is already familiar to you jump ahead to the second to last section titled Using the Taxon Split input as the output.
Taxon Swaps
On iNaturalist if you want to change the main scientific name associated with a taxon (e.g. from Species X to Species Y) you do the following steps:
- Create a new inactive taxon with the name Species Y
- Create a draft Taxon Swap with Species X as the input and Species Y as the output
- Commit the Taxon Swap
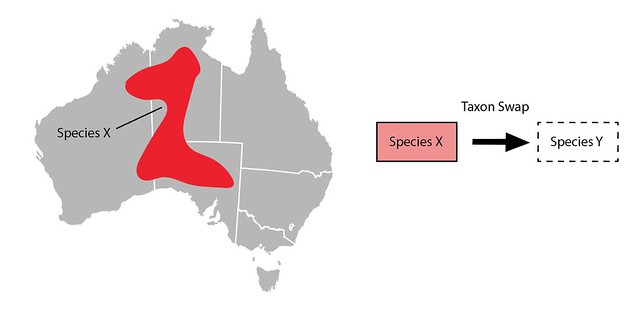
Why do all this instead of just changing the name on the taxon? Just changing the name would mean that if an identifier typed in an identification of Species X it might unexpectedly change to Species Y without any record of what happened. Create a taxon swap leaves a record so identifiers can see when, how, and why the name associated with their identification changed.
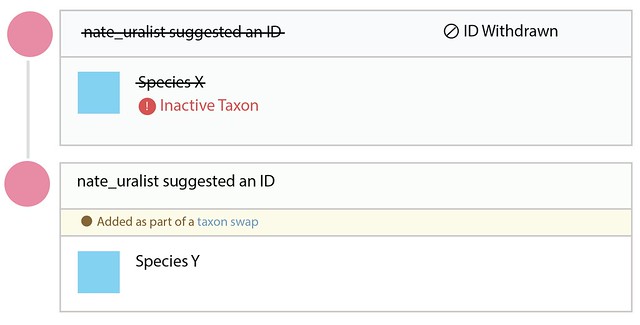
Let’s call this updating identifications. We’re aware that this might not be the easiest, least disruptive way to generate such a log, especially when the name changes are trivial such as going from Cnemaspis hitihami to Cnemaspis hitihamii. But under the current functionality, taxon swaps are always used to change the main scientific name of a taxon.
As shown above, taxon swaps have a single output which means all identifications of the input taxon can be unambiguously updated with identifications of the output taxon. Likewise, other content associated with the input taxon, such as taxon names (where iNaturalist stores common names and synonyms), taxon ranges, listed taxa etc., is moved or copied over to the output taxon*. Committing a taxon swap also makes the input taxon inactive and activates inactive output taxa.
Using Taxon Swaps to Lump Taxa
Another common curation action is to lump one taxon into another (e.g. Species X into Species Z). A taxon swap can also be used to do this by making Species X the input and the already active Species Z as the output. As described above, committing the taxon swap will inactivate Species X, update all identifications, and copy/move other content over to Species Z.
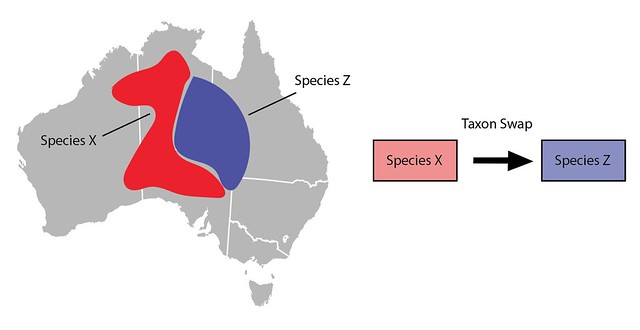
One thing to keep in mind when using Taxon Swaps to lump taxa is that lumping often broadens what we mean by the output taxon. In this example Species Z’s taxon range to the east no longer matches its newly broadened meaning and would need to be manually modified to include the western part of the range. Similarly, imagine Species X’s common name was "Western Critter" and Species Z’s common name was "Eastern Critter". Post-swap, Species’ Z’s common name might need to be updated to something like "Critter".

Several uses for Taxon Merges
If you want to lump two or more taxa (Species X and Species Q) into an existing taxon (Species Z) you can use a Taxon Merge. Taxon Merges work like taxon swaps but have multiple input taxa. Since there’s just a single output taxon, identifications and content are also migrated to the output taxon*.
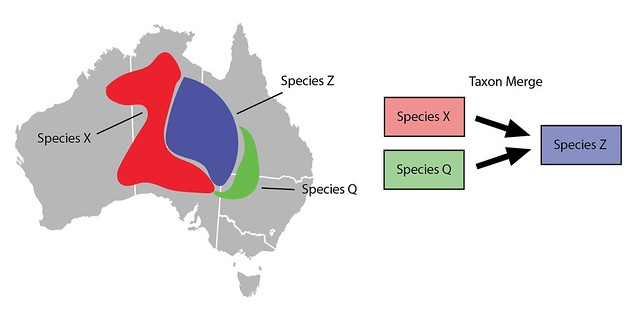
Taxon Merges can also be used to lump taxa slightly differently. Imagine that rather than manually broadening all the content associated with Species Z you prefer to just make a new taxon for Species Z. Let's call it Species Z’. It would start out inactive, would have the same main scientific name as Species Z, but would have a different broader meaning and thus different broader content such as range maps.
Committing the Taxon Merge would inactivate the input Species Z and activate the broader output Species Z', identifications of Species Z would be updated with identifications of Species Z’ and all other content would be copied to Species Z’.
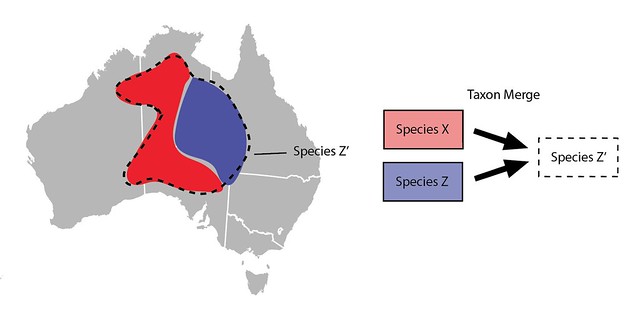
This approach requires more steps than just manually broadening Species Z (Species Z’ must be created etc.) and perhaps unnecessarily updates all the identifications of Species Z with Species Z` (unnecessary since both taxa have the same the main scientific name). So this way of lumping taxa is generally not preferred, but it is an option available to curators.
Taxon Splits
To carve one taxon off from another (e.g. Species W off from Species X) we use Taxon Splits. Taxon splits have a single input and more than one output. The only way to make taxon splits in the past was to create new inactive taxa for the carved-off taxon (Species W) and for the new narrowed version of the input taxon (Species X’).

Since splits have more than one output taxon, identifications are updated to the common ancestor of the outputs. Other content such as range maps and names are not copied over to the output taxa and must be added manually*.

Atlases provide some flexibility for determining a single output taxon with which to update identifications. Using the example of the atlases below, identifications associated with observations in Western Australia (WA) and South Australia (SA) would be updated with output taxon Species X’ while identifications belonging to observations in Queensland (QLD) would be updated with output taxon Species W. Only identifications belonging to observations within presence places in both atlases (e.g. Northern Territory, NT) or outside of both atlases (e.g. New South Wales) would be updated with the common ancestor.

Using the Taxon Split input as the output
Sometimes only a tiny piece of a taxon is carved off as a new taxon. When the input taxon has a lot of identifications this cran result in many unecessary identification updates (e.g. Species X with Species X’).
We’ve made some changes that allow curators the flexibility, if they choose, to make Taxon Splits where the input taxon is also one of the output taxa. In some ways, this approach is analogous to using a taxon swap to broaden an existing taxon (e.g. Species X) instead of using a taxon merge with a new output taxon (e.g. Species X’) as was described above. In this approach, the input taxon is first manually narrowed by modifying the atlas (as well as other content such as range map, etc.) which will be used to determine the destination of identifications.

Upon committing such a Taxon Split, iNaturalist will not inactivate the input taxon. It will still use the atlases to determine which identifications should be updated with which output taxon, but any identification determined to be updated with the output taxon matching the input (e.g. Species X) will not be touched.
Because doing a split this way won’t update as many identifications (e.g. updating identifications of Species X with identifications of Species X’), it will always be less disruptive. But it’s important to remember to manually narrow the input taxon by updating content such as the taxon range and common names that likely no longer match the narrowed meaning taxon after the split.
There are some advantages from the normal way of splitting taxa such as having the inactivated input taxon (Species X) persist alongside the narrowed taxon (Species X’) coexisting in the database for a more thorough comparison of how the taxon was narrowed. However, as iNaturalist has grown, it is our feeling that the disruptive costs both in terms of processing time and in terms of identification clutter of having so many updated identifications (e.g. Species X to Species X’) now outweighs these advantages, especially when there are a lot of identifications on the input taxon (Species X) that are not destined to be carved off (e.g. as Species W). As long as curators remember to manually narrow content such as taxon ranges, we anticipate that this new way of splitting taxa will generally be the preferred way to split taxa moving forward.
Examples
Here are two examples of this new kind of taxon split I committed. The first involved splitting two Caribbean island endemics (Boa orophias on St. Lucia and, Boa nebulosa on Dominica) off from Boa constrictor. Notice that the same Boa constrictor taxon is listed as both the input and an output.

As expected, this observation from St. Lucia was properly updated to Boa orophias whereas this captive observation outside of all three atlases was updated to Genus Boa. And this observation from within the range of Boa constrictor was untouched.
Similarly, this split involved splitting two species (Varanus tsukamotoi from Guam and other Pacific Islands and Varanus bennetti from several other islands including Palau) off from Varanus indicus. Again note that the same taxon Varanus indicus acts as both input and output.

As expected, this observation from Palau was updated as Varanus bennetti, this observation from Guam was updated as Varanus tsukamotoi, and this observation from Papua was untouched.
Note that in both of these examples, I manually narrowed both the taxon range and atlas of the input taxa before committing the splits.
*The details about which content is copied over and which is moved over to the output taxon when a taxon change is committed is complicated. Here’s an attempt to explain exactly what happens. In both a taxon swap and a taxon merge, taxon photos and taxon names are copied over. The copy of the valid scientific name from the input taxon is invalid (a synonym) on the output taxon. Taxon swaps, but not taxon merges, copy the taxon range, conservation statuses, and atlas from the input (unless a taxon range or atlas or conservation status for the same place already exists on the output taxon). Listed taxa are moved/merged from the input taxon to the output taxon for both taxon swaps and taxon merges. For taxon splits, no content is copied over (since the output taxon is ambiguous). Listed taxa are moved to a single output taxon if atlases can be used to uniquely determine one, much like atlases are used to update identifications.